Arby Abood
PhD candidate
University of Virginia
Center for Public Health Genomics
Data Science Training Program
Bio
I’m a PhD candidate, bioinformatician and data scientist at The University of Virginia School of Medicine. My research focuses on identifying theraputic targets for ostoeoporosis using systems genetics. I am currently integrating genome-wide assocition studies, splice quantitative trait loci, long-read RNA sequencing, and proteomics to identify novel isoforms as candidate causal mediators of disease.
Interests
- Systems Genetics
- Computational Biology
- Genetic Epidemiology
Education
PhD in Genomics & Bioinformatics, 2023
University of Virginia
MSc in Microbial Genomics, 2018
Clemson University
BSc in Biology, 2013
George Mason University
Skills
R
Python
Genetic epidemiology and statistics
Bash
Experience
PhD candidate
University of Virginia
Responsibilities include:
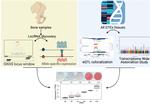
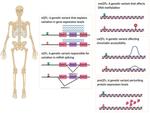
- Informed bone mineral density (BMD) genome-wide association studies (GWAS) by integrating splice quantitative trait loci (sQTL) data with Bayesian colocalization analysis. The results were further refined by using data from long-read RNAseq, and proteomics followed by experimental validation in-vitro. This enabled us to implicate causal genes in osteoporosis.
- Systematically identified long non-coding RNAs potentially responsible for the effect of BMD GWAS loci using expression quantitative trait loci (eQTL) Bayesian colocalization, transcriptome-wide association studies (TWAS), and allelic imbalance analyses. Due to this work, we were able to shed light on the non-coding aspects of the bone transcriptome and their role in osteoporosis.
- Characterized the landscape of isoforms and proteoforms in the process of osteoblast differentiation by combining long-read RNAseq data and proteomics data and applying differential expression analysis, differential splicing analysis, differential isoform usage, and other statistical approaches.
Graduate assistant
Clemson University
- Investigated human oral and gut microbiome community dynamics using both 16S and Whole Genome Shotgun (WGS) metagenomics data analyses. This enabled us to understand host-microbiome interaction and provide detailed abundance counts at bacterial strain level resolution.
- Implemented multiple bioinformatics pipelines and provided detailed instructions on their usage to assist lab partners and collaborators with their research needs.
- Managed day-to-day laboratory operations including ordering, and Linux server maintenance.
- Gained experience in the bash/Linux command line interface (CLI), Python, and R.
Accomplishments
Become a Python Developer
Data scientist & analyst with Python
Data scientist & analyst with R
Projects
other
An example of using the in-built project page.
Transcriptomics
An example of using the in-built project page.